AI-Designed Drugs & Biologics: Accelerating the Drug Discovery Curve
Artificial intelligence is redefining drug discovery, driving a shift from small molecules to biologics and enabling the precise design of advanced therapeutics for complex diseases.

|
October 15, 2025
|
9 min read
The AI Revolution in Drug Discovery
Artificial intelligence is transforming drug discovery at an unprecedented pace, opening new possibilities for both small molecules and biologics. While most AI platforms have traditionally focused on small molecule design, there's a growing shift toward biologics; complex, highly specific drugs that are changing the treatment landscape for oncology, immunology, and rare diseases. As AI-generated molecules become more popular and companies increasingly outsource their laboratory work, the importance of expertise and validation in this space cannot be overstated.
From Small Molecules to Biologics: Why the Shift Matters
Historically, small molecules have dominated the pharmaceutical market due to their oral bioavailability. This simplifies early-stage pharmacokinetic profiling by enabling straightforward assessment of ADME (Absorption, Distribution, Metabolism, Excretion) characteristics. Small molecule drugs typically follow a well-characterised absorption route:
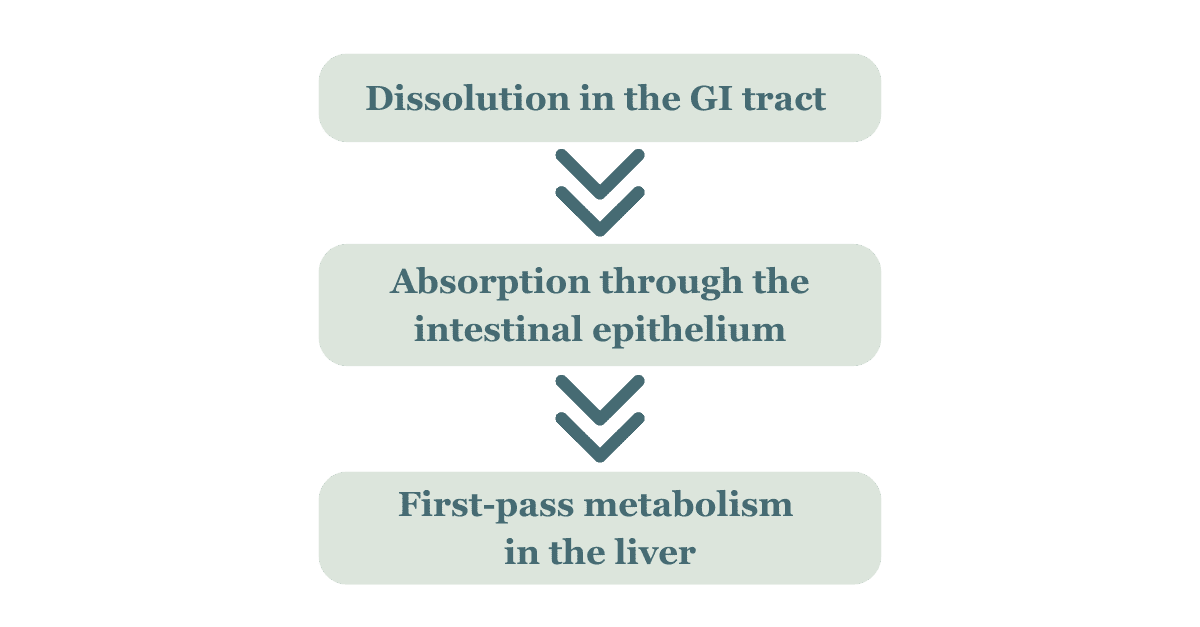
This predictability allows researchers to model absorption kinetics more easily, identify any rate-limiting steps such as solubility or permeability, and to use established in vitro and in vivo models to stimulate human pharmacokinetic behaviour. Small molecules also have physiochemical properties that make them ideal for efficient and predictable distribution, such as high tissue penetration, favourable volume of distribution, predictable plasma protein binding and compatibility with oral dosing.
Metabolism in small molecules is favourable in the pharmaceutical market because it is predictable, well-studied, and easily optimised. Researchers can adjust molecular structures to improve metabolic stability, reduce toxicity, or extend half-life. Some small molecules even produce active metabolites that enhance therapeutic effects. Excretion in small molecules is favourable in the pharmaceutical market because it is efficient, predictable, and easy to assess. These compounds are typically small and polar, allowing for straightforward elimination through urine or bile. Their well-characterised excretion pathways reduce the risk of accumulation and toxicity, especially in long-term treatments.
AI-Driven Drug Discovery: Adapting to Complexity
Applying AI in small molecule drug design can accelerate discovery, reduce costs, and improve precision. It enables rapid screening of thousands of compounds, predicting which are most likely to succeed based on ADME properties. Small molecules are also favoured in AI drug design as they have a simplified manufacturing process. This allows for streamlined development and testing, due to the fact that they can be administered non-invasively, thereby reducing the need for complex delivery systems.
As AI tools become more integrated across the drug development pipeline, they are reshaping how researchers identify, validate and refine therapeutics. AI has been widely adopted for small molecule discovery, where it has accelerated lead identification, optimised compound properties, and reduced development timelines. But as biologics prove to be better suited for intricate targets like protein-protein interactions (PPIs), the use of AI is quickly expanding to these larger, more complex drugs.
Does AI Overcome Biologics Design Challenges?
Biologics are uniquely capable of targeting PPIs, which are often inaccessible to small molecules due to their flat or diffuse binding surfaces. However, their structural complexity and sensitivity to environmental conditions present significant challenges in design, optimisation, and validation.
AI is helping overcome these hurdles by:
- Predicting protein structure and folding with high accuracy using models like AlphaFold and EvoFormer.
- Characterising higher-order structures and glycosylation patterns, which are critical for biologic function and safety.
- Designing mechanism-reflective assays that simulate real biological conditions, enabling more precise evaluation of biologic candidates.
This convergence of biologics and AI is not just a technical upgrade; it’s a key shift in how we approach drug design for complex diseases. PPIs are central to cellular function, yet notoriously difficult to model due to their dynamic and context-dependent nature. Deep learning is now transforming this space.
Recent advances include transformer-based models and graph neural networks that integrate sequence and structural data to predict PPIs with unprecedented accuracy, as well as multimodal frameworks that combine omics data, enabling AI to capture the full biological context of interactions. These innovations are not just improving prediction; they’re enabling the design of biologics that can modulate PPIs with optimal precision.
How does AI Push the Boundaries in Drug Discovery Now?
AI is no longer just a tool for discovery, it’s becoming a co-creator in biologic design, particularly in the development of antibody therapeutics. This shift is evident in several key innovations that are reshaping how biologics are conceived and refined.
One major advancement is AI-guided fragment design, where platforms like FragFold predict short protein sequences that will be able to inhibit target proteins by mimicking native interactions. These fragments offer a modular and precise approach to biologic development, especially for challenging targets like PPIs.
In parallel, generative AI platforms, such as those developed by LabGenius, Absci, and Generate:Biomedicines, are being used to create novel antibody formats. These models optimise critical features like binding affinity, molecular stability, and immunogenicity, often outperforming traditional design methods in speed and precision.
Further pushing the boundaries, closed-loop design systems like AuraQuest and AuraGlow integrate real-time experimental data with predictive modelling. This iterative approach allows biologic candidates to be refined continuously, enhancing their therapeutic potential while reducing development cycles.
With AI now embedded in the design loop, biologics are evolving faster, smarter, and with greater therapeutic precision than ever before. The move toward biologics is not just about innovation; it's about specificity, precision, and the opportunity to tackle conditions once deemed untreatable. It’s important, however, to retain balanced positioning: most AI efforts still centre on small molecules, and the market remains primarily there. Yet, with biologics becoming increasingly relatable and relevant, the potential for AI-driven advances in this domain is enormous.

Designing Antibodies with AI: Platforms, Methods, and Breakthroughs
In recent years, AI platforms have begun designing antibodies de novo, crafting all complementarity-determining regions with binding affinities that surpass those achieved through random sampling. This represents a significant leap in biologic design, where precision and specificity are paramount. Companies like Absci have demonstrated generative models capable of producing binders with lower affinities than trastuzumab, one of the most widely used therapeutic antibodies on the market. These AI-driven approaches are not only accelerating discovery but also improving the quality of candidates entering preclinical pipelines.
While the promise of AI in antibody design is undeniable, it’s important to maintain a balanced perspective. A recent open AI competition, led by Andrew Bradbury of Specifica, challenged AI models to design de novo antibodies for a well-studied target. Although expectations were high, the results did not fully live up to the initial hype, underscoring that the field still has a long way to go before AI can consistently deliver breakthrough biologics. This example highlights the need for continued innovation, rigorous validation, and realistic expectations as AI technologies evolve.
Despite these challenges, the innovation doesn’t stop at single antibodies. AI is now driving the development of multispecific antibodies, antibody-drug conjugates, and engineered Fc regions, each offering enhanced therapeutic potential through improved targeting, immune modulation, and pharmacokinetics. Leaders in this space are redefining biologic development by optimising key attributes such as developability, immunogenicity, and binding affinity, all within integrated computational frameworks.
Wet Lab Validation: Why AI Still Needs the Bench
Despite AI’s remarkable predictive capabilities, wet lab validation remains critical. While AI can simulate molecular interactions, predict binding affinities, and optimise therapeutic properties, it cannot fully replicate the complexity of biological systems.
Biophysical techniques such as surface plasmon resonance (SPR) and biolayer interferometry (BLI) offer real-world confirmation of the binding kinetics of an interaction between antigen (or Fc Receptor) and antibody. While cell-based bioassays provide empirical validation of the expected biological activity of an antibody, and can include mechanism of action determination, using an appropriate model, but also extend to antibody effector function assays such as antibody dependent cell mediated cytotoxicity (ADCC), complement dependent cytotoxicity (CDC) and antibody dependent cellular phagocytosis (ADCP). Having their place among other product characterisation assays, determination of function and binding characteristics remain critical for efficacy.
Another key consideration is that novel, non-native formats may increase the risk of immunogenicity. AI-designed sequences could present epitopes that humans have never been exposed to, raising the likelihood of immune recognition and strong antibody responses. This in turn can lead to unintended outcomes such as cytokine storms from immune over-activation or the inadvertent introduction of allergenic motifs. To mitigate these risks, immunotoxicological assessments are essential - including cytokine release assays, measurement of T cell responses, and monitoring of pre-existing anti-drug antibodies (ADAs), or treatment-induced ADAs. Together, these evaluations help ensure that AI-generated molecules behave as intended under physiological conditions and satisfy regulatory safety standards.
Regulatory agencies increasingly demand contextually relevant, reproducible, and validated data, and AI-only workflows often fall short without iterative feedback from experimental results. This makes wet lab validation not just a formality, but a strategic necessity in the development pipeline.
Our Immunology CRO services are built around validating both AI-derived and traditional molecules. The difference rests only in the source of the model; our workflow brings the same rigour and quality to both, but with the added advantage of higher throughput, screening thousands of AI-generated candidates to find the most promising therapeutic.
Challenges and Ethical Considerations
As AI becomes increasingly integral to drug design, it brings both transformative potential and complex challenges. While AI can enhance accuracy by drawing from vast datasets, the current landscape is hindered by limited, inconsistent, and often low-quality data. This affects not only prediction reliability but also transparency, especially when models generate outputs not grounded in real evidence, commonly referred to as AI hallucinations. To mitigate this, the use of high-quality, diverse datasets is essential.
Bias is another critical issue. AI systems trained on unbalanced or incomplete data may inherit and amplify existing biases, leading to skewed results and reduced trust. Ethical concerns also arise, particularly around accountability, especially when AI is used to inform decisions about human health. Ensuring that AI systems are transparent and interpretable is vital for their acceptance by healthcare professionals.
Data privacy and security are equally important. AI-driven drug discovery often involves sensitive patient and clinical data, which must be protected to comply with regulations and prevent unauthorised access. Compounding these challenges is the lack of clear regulatory frameworks. As AI is still a relatively new tool in drug development, policies are evolving, and this uncertainty can complicate implementation.
Another practical challenge is the gap in biological expertise among many AI companies. While they excel in computational modelling, they often lack the in-house knowledge needed to validate and develop molecules effectively. This is where partnerships with CROs (like RoukenBio) become invaluable; not only do they provide wet-lab facilities, but they also offer experience, scientific leadership and guidance, helping AI drug developers navigate validation, regulatory expectations, and ethical standards.
Despite these hurdles, AI is reshaping the drug discovery landscape. Researchers and developers must adapt to this shift, leveraging partnerships and robust data practices to optimise time, resources, and future patient outcomes.
Future Outlook: Autonomous Drug Design and Personalised Biologics
The future of drug discovery lies in the synergy between AI and wet lab expertise. Autonomous drug design is on the horizon, promising personalised biologics tailored to individual patients. By leveraging patient-specific data such as genomic sequences, proteomic signatures, and immune markers, AI has the potential to design biologics that offer higher efficacy, reduced side effects, and improved long-term outcomes. As AI technologies mature, the role of R&D partners with a deep understanding of both computation and experimentation will be more important than ever.
The ability to translate digital designs into viable, scalable treatments will define the next generation of drug development. In this evolving landscape, organisations that combine high-throughput lab capabilities, biological insight, and AI fluency will be best positioned to lead. The shift toward autonomous and personalised therapeutics is not just a technological evolution; it’s a strategic transformation in how we approach human health.
Join our community of curious minds on LinkedIn
🗓️ Stay informed with our monthly scientific newsletter, published on LinkedIn on the last Wednesday of each month.
These editions bring you the latest in drug development breakthroughs, industry trends, and expert insights from the brilliant minds at RoukenBio.
Subscribe today on LinkedInValidate with RoukenBio
Our laboratory facilities and biological expertise set us apart to support AI companies from screening of initial computational designs through successive iterations, to rigorous experimental validation. As AI-designed products gain momentum the industry moves rapidly towards outsourcing, we stand ready as your partner CRO to help you triage your potential candidates into a single, game-changing therapeutic.
Download our Capabilities Brochure

